Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology* IBMC-CNRS, Universite Louis Pasteur**
Yoshiteru Sato* Kenta Mitomi* Hiromu Sugiyama* Satoru Shimizu* Ella Czarina Magat Juan* Jiro Kondo** ○Akio Takenaka*
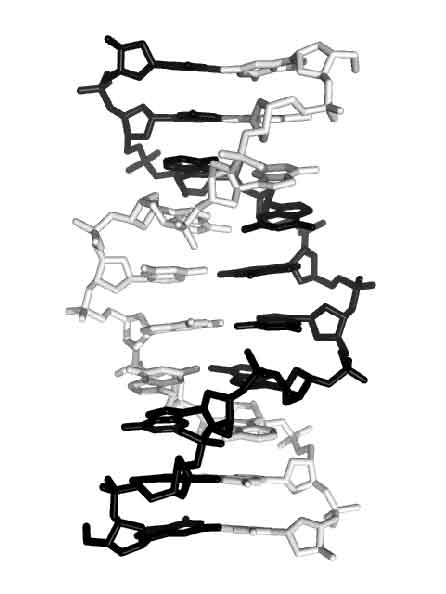
We found that DNA fragments with the sequence d(gcGA[X]1Agc) (X=G and A) form an base-intercalated duplex and that it is the basic unit for further assembly to form an octaplex. To investigate versatility of multiplex formation, d(gcGA[G]3Agc) was crystallized in the two forms: P212121 and P21. The crystal structure of the latter form has been determined at 1.8 Å resolution. The two DNA fragments in the asymmetric unit form a duplex. The most interesting finding is that two sheared-type G3:A8 and A4:G7 pairs, two G5:G6 and G6:G5 pairs and two sheared-type G7:A4 and A8:G3 pairs are formed in the major part, which suggests that the duplex is formed by non-WC pairings, though two WC type G1:C10 and C2:G9 pairs occur at both ends (see the figure). Native gel electrophoresis experiments, performed to examine such a possibility, shows that all DNA fragments with d(GAG[G]nGA) (where n=1-4) form not only duplexes, but also several multiplexes such as quadruplexes, octaplexes, and so on. In human genome, these types of sequences are frequently found in SNPs of purine rich regions. They might have some functions with specific structures. These sequences are crystallized.

Fig. Overall duplex structure of d(gcGA[G]3Agc)